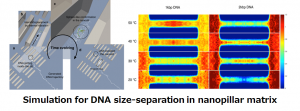
 The aim of this research is to establish microfluidic system to perform size-separation of biomolecules such as DNA and DNA nanostructures having different size and shapes and computational tool to assist the design process for the system.
The aim of this research is to establish microfluidic system to perform size-separation of biomolecules such as DNA and DNA nanostructures having different size and shapes and computational tool to assist the design process for the system.
[Applications]
- Purification of DNA origami after fabrication
- Size-separation of polymerized DNA origami tiles
[Publications]
- A Constrained Rigid Body Brownian Dynamics Simulation for On-Chip Size-Separation of Cross-linked DNA Origami Tiles, 28th International Microprocesses and Nanotechnology Conference, Toyama Japan, November 2015.
- Brownian Dynamics Simulation for Electrophoretic Migration of DNA Nanostructures in a 2D Anisotropic Nanosieving Structure, The Institute of Electrical Engineers of Japan (IEEJ) Sensor/Micromachine Division Symposium, Hakata, Kyushu, Japan, 2 – 3 July 2015
- A Multibody Brownian Dynamics for Modeling Size-separation of dsDNA Fragment and DNA Origami Tiles in Anisotropic Nanosieving Array, NEMS 2016 (Accepted)